-Search query
-Search result
Showing 1 - 50 of 670 items for (author: paul & ss)

EMDB-17375: 
Neisseria meningitidis Type IV pilus SB-GATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17384: 
Neisseria meningitidis Type IV pilus SB-DATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17386: 
Neisseria meningitidis Type IV pilus SA-GATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17683: 
Neisseria meningitidis Type IV pilus SB-GATDH variant bound to the C24 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17695: 
Neisseria meningitidis Type IV pilus SB-DATDH variant bound to the C24 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17718: 
Neisseria meningitidis PilE, SB-GATDH variant, bound to the F10 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8p2v: 
Neisseria meningitidis Type IV pilus SB-GATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8p36: 
Neisseria meningitidis Type IV pilus SB-DATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8p3b: 
Neisseria meningitidis Type IV pilus SA-GATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8pij: 
Neisseria meningitidis Type IV pilus SB-GATDH variant bound to the C24 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8piz: 
Neisseria meningitidis Type IV pilus SB-DATDH variant bound to the C24 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8pjp: 
Neisseria meningitidis PilE, SB-GATDH variant, bound to the F10 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-43658: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-43659: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-43660: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vye: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vyf: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vyg: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-40468: 
In situ human cardiac thick filament in the relaxed state
Method: subtomogram averaging / : Chen L, Liu J, Rastegarpouyani H, Janssen PML, Pinto JR, Taylor KA

EMDB-40471: 
Human cardiac interacting heads motif (IHM-C) in complete form
Method: subtomogram averaging / : Chen L, Liu J, Rastegarpouyani H, Janssen PML, Pinto JR, Taylor KA

EMDB-40475: 
Human cardiac interacting heads motif (IHM-C) in semi form
Method: subtomogram averaging / : Chen L, Liu J, Rastegarpouyani H, Janssen PML, Pinto JR, Taylor KA

EMDB-40476: 
Human cardiac interacting heads motif (IHM-S)
Method: subtomogram averaging / : Chen L, Liu J, Rastegarpouyani H, Janssen PML, Pinto JR, Taylor KA

EMDB-40478: 
Human cardiac interacting heads motif (IHM-D) in semi form
Method: subtomogram averaging / : Chen L, Liu J, Rastegarpouyani H, Janssen PML, Pinto JR, Taylor KA

EMDB-16933: 
Structure of cGAS in complex with SPSB3-ELOBC
Method: single particle / : Xu PB, Ablasser A

EMDB-16936: 
cGAS-Nucleosome in complex with SPSB3-ELOBC (composite structure)
Method: single particle / : Xu PB, Ablasser A

EMDB-16937: 
Consensus refinement of cGAS/spsb3/EloBC/Nucleosome
Method: single particle / : Xu PB, Ablasser A
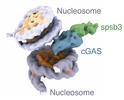
EMDB-16938: 
human cGAS/spsb3/EloBC in complex with nucleosome (2:2)
Method: single particle / : Xu PB, Ablasser A

PDB-8okx: 
Structure of cGAS in complex with SPSB3-ELOBC
Method: single particle / : Xu PB, Ablasser A

PDB-8ol1: 
cGAS-Nucleosome in complex with SPSB3-ELOBC (composite structure)
Method: single particle / : Xu PB, Ablasser A

EMDB-19194: 
In situ cryo-electron tomogram of an autophagosome in the projection of an iPSC-derived neuron #2
Method: electron tomography / : Hoyer MJ, Capitanio C, Smith IR, Paoli JC, Bieber A, Jiang Y, Paulo JA, Gonzalez-Lozano MA, Baumeister W, Wilfling F, Schulman BA, Harper WJ

EMDB-19346: 
In situ cryo-electron tomogram of an autophagosome in the projection of an iPSC-derived neuron #1
Method: electron tomography / : Hoyer MJ, Capitanio C, Smith IR, Paoli JC, Bieber A, Jiang Y, Paulo JA, Gonzalez-Lozano MA, Baumeister W, Wilfling F, Schulman BA, Harper WJ

EMDB-18245: 
Plunge-frozen (control) map of beta-galactosidase
Method: single particle / : Esser TK, Boehning J, Bharat TAM, Rauschenbach S

EMDB-17659: 
ACAD9-WT in complex with ECSIT-CTER
Method: single particle / : McGregor L, Acajjaoui S, Desfosses A, Saidi M, Bacia-Verloop M, Schwarz JJ, Juyoux P, Von Velsen J, Bowler MW, McCarthy A, Kandiah E, Gutsche I, Soler-Lopez M

EMDB-17660: 
Cryo-EM structure of human ACAD9-S191A
Method: single particle / : McGregor L, Acajjaoui S, Desfosses A, Saidi M, Bacia-Verloop M, Schwarz JJ, Juyoux P, Von Velsen J, Bowler MW, McCarthy A, Kandiah E, Gutsche I, Soler-Lopez M

EMDB-17661: 
ACAD9 homodimer WT
Method: single particle / : McGregor L, Acajjaoui S, Desfosses A, Saidi M, Bacia-Verloop M, Schwarz JJ, Juyoux P, Von Velsen J, Bowler MW, McCarthy A, Kandiah E, Gutsche I, Soler-Lopez M

PDB-8phe: 
ACAD9-WT in complex with ECSIT-CTER
Method: single particle / : McGregor L, Acajjaoui S, Desfosses A, Saidi M, Bacia-Verloop M, Schwarz JJ, Juyoux P, Von Velsen J, Bowler MW, McCarthy A, Kandiah E, Gutsche I, Soler-Lopez M

PDB-8phf: 
Cryo-EM structure of human ACAD9-S191A
Method: single particle / : McGregor L, Acajjaoui S, Desfosses A, Saidi M, Bacia-Verloop M, Schwarz JJ, Juyoux P, Von Velsen J, Bowler MW, McCarthy A, Kandiah E, Gutsche I, Soler-Lopez M

EMDB-18244: 
ESIBD structure of beta-galactosidase
Method: single particle / : Esser T, Boehning J, Bharat TAM, Rauschenbach S

PDB-8q7y: 
ESIBD structure of beta-galactosidase
Method: single particle / : Esser T, Boehning J, Bharat TAM, Rauschenbach S

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
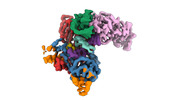
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
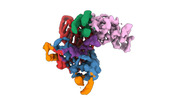
EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxt: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxu: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

EMDB-27641: 
The structure of the interleukin 11 signalling complex, truncated gp130
Method: single particle / : Metcalfe RD, Hanssen E, Griffin MDW

EMDB-27642: 
The structure of the IL-11 signalling complex, with full-length extracellular gp130
Method: single particle / : Metcalfe RD, Hanssen E, Griffin MDW

PDB-8dps: 
The structure of the interleukin 11 signalling complex, truncated gp130
Method: single particle / : Metcalfe RD, Hanssen E, Griffin MDW

PDB-8dpt: 
The structure of the IL-11 signalling complex, with full-length extracellular gp130
Method: single particle / : Metcalfe RD, Hanssen E, Griffin MDW
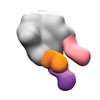
EMDB-40242: 
BG505 MD39 SOSIP in complex with Rh.NJ85 wk12 gp120GH, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB

EMDB-40243: 
BG505 MD39 SOSIP in complex with Rh.NJ86 wk12 V1V3, C3V5, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
